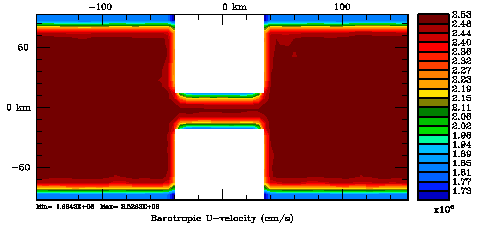
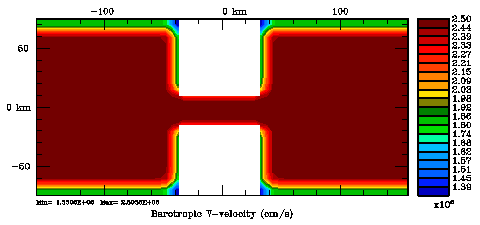
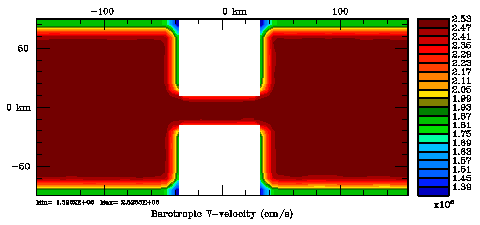
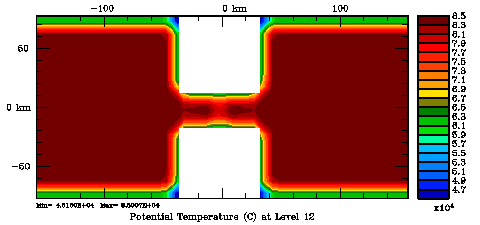
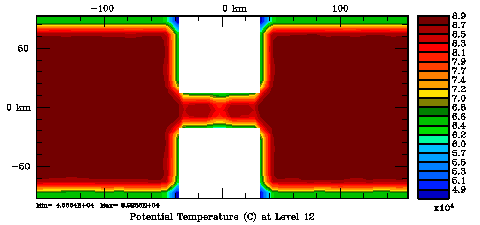
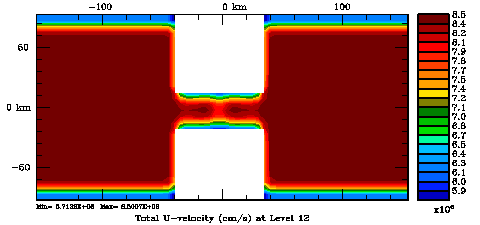
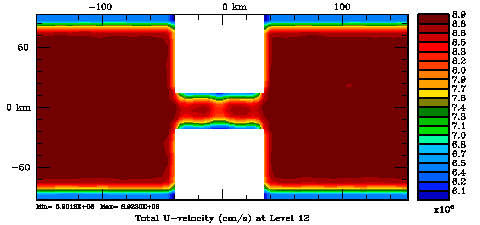
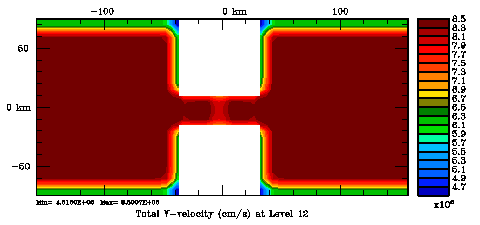
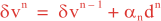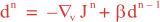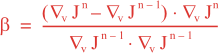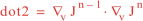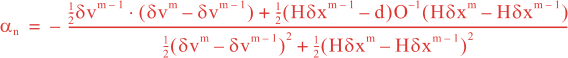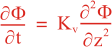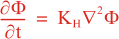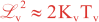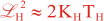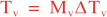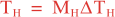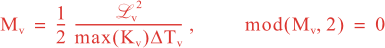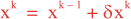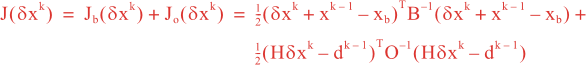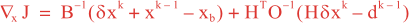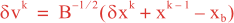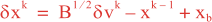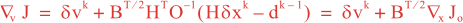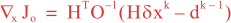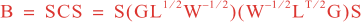Following Weaver and Courtier (2001), the vertical and horizontal correlations, C, for any state variable Φ are modeled using separated 1D and 2D diffusion equations:
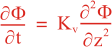
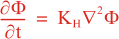
where Kv and KH are the vertical and horizontal diffusion coefficients (m2/s), assumed to be of order one. They are set to unity (isotropic correlations), if these coefficients are not found in ROMS/TOMS background/model standard deviation NetCDF file (STDname, variables Kv and Kh). The user can set Kv and Kh to the desired patterns to get anisotropic correlations. If we discretize the above diffusion operators using an explicit FTCS scheme, the stability limit is governed by:


In practice, we set γ < 1 to be below the stability threshold, (γ = 0.5). This stability parameter is set in 4DVAR input script. There are two variables: Vgamma and Hgamma for vertical and horizontal diffusion operators. In realistic applications, the vertical difussion needs to be solved implicitly (IMPLICIT_VCONV). Otherwise, the it will be very expensive since the vertical scales are much smaller. The implicit algorithm is unconditionally stable for any time-step, so we use the maximum thickness in the above equation instead of the minimum.
The parameters controlling the vertical and horizontal length-scales of the Gaussian correlation function are:
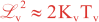
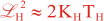
over a time interval 0 ≤ t ≤ T and
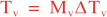
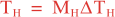
where Mv and MH are the total number of vertical and horizontal integration steps. Notice that both values are forced to an even number since the squared-root diffusion operators are used to insure symmetry. The squared-root operator iterates only for half of the steps. For any given decorrelation length-scale, the number of integration steps are given by:
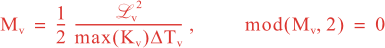

The vertical and horizontal decorrelation scales (m) are set in 4DVar input script variables Vdecay(:) and Hdecay(:) for each state variable.
ROMS/TOMS reports to standard output all the parameters associated with these diffusion operators. For example,
5.0000E-01 Hgamma Horizontal diffusion stability/accuracy factor.
5.0000E-05 Vgamma Vertical diffusion stability/accuracy factor.
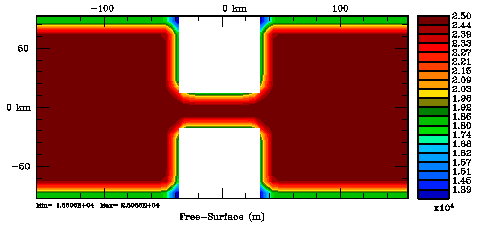
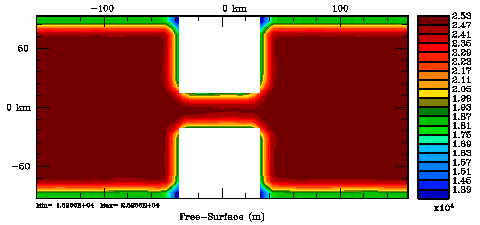
20000.000 Hdecay(isFsur) Horizontal decorrelation scale (m), free-sruface.
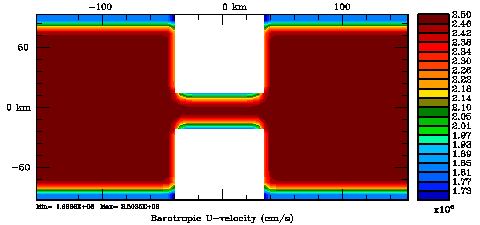
20000.000 Hdecay(isUbar) Horizontal decorrelation scale (m), 2D U-momentum.
20000.000 Hdecay(isVbar) Horizontal decorrelation scale (m), 2D V-momentum.
20000.000 Hdecay(isUvel) Horizontal decorrelation scale (m), 3D U-momentum.
20000.000 Hdecay(isVvel) Horizontal decorrelation scale (m), 3D V-momentum.
20000.000 Hdecay(idTvar) Horizontal decorrelation scale (m), temp.
20000.000 Hdecay(idTvar) Horizontal decorrelation scale (m), salt.
5.000 Vdecay(isUvel) Vertical decorrelation scale (m), 3D U-momentum.
5.000 Vdecay(isVvel) Vertical decorrelation scale (m), 3D V-momentum.
5.000 Vdecay(idTvar) Vertical decorrelation scale (m), temp.
5.000 Vdecay(idTvar) Vertical decorrelation scale (m), salt.
Minimum X-grid spacing, DXmin = 5.26835784E+00 km
Maximum X-grid spacing, DXmax = 5.58390264E+00 km
Minimum Y-grid spacing, DYmin = 5.05224079E+00 km
Maximum Y-grid spacing, DYmax = 5.35460591E+00 km
Minimum Z-grid spacing, DZmin = 1.33333333E-01 m
Maximum Z-grid spacing, DZmax = 2.86550363E+02 m
Minimum barotropic Courant Number = 2.99123135E-02
Maximum barotropic Courant Number = 8.17259140E-01
Maximum Coriolis Courant Number = 3.43845829E-02
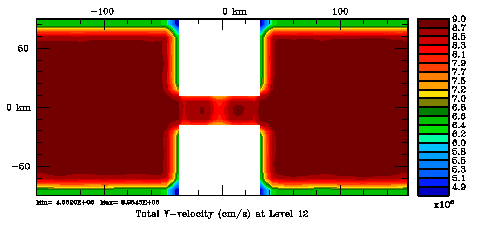
Horizontal convolution, NHsteps, DTsizeH = 64 3.19064213E+06 s zeta
Horizontal convolution, NHsteps, DTsizeH = 64 3.19064213E+06 s ubar
Horizontal convolution, NHsteps, DTsizeH = 64 3.19064213E+06 s vbar
Horizontal convolution, NHsteps, DTsizeH = 64 3.19064213E+06 s u
Horizontal convolution, NHsteps, DTsizeH = 64 3.19064213E+06 s v
Horizontal convolution, NHsteps, DTsizeH = 64 3.19064213E+06 s temp
Horizontal convolution, NHsteps, DTsizeH = 64 3.19064213E+06 s salt
Vertical convolution, NVsteps, DTsizeV = 6 2.05277776E+00 s u
Vertical convolution, NVsteps, DTsizeV = 6 2.05277776E+00 s v
Vertical convolution, NVsteps, DTsizeV = 6 2.05277776E+00 s temp
Vertical convolution, NVsteps, DTsizeV = 6 2.05277776E+00 s salt
It is recommended to reduce the Vgamma parameter until a value of Mv is around six for vertical diffusion accuracy.