#865 closed upgrade (Done)
IMPORTANT: Split 4D-Var algorithms, Phase II
| Reported by: | arango | Owned by: | |
|---|---|---|---|
| Priority: | major | Milestone: | Release ROMS/TOMS 3.9 |
| Component: | Nonlinear | Version: | 3.9 |
| Keywords: | Cc: |
Description (last modified by )
Testing is completed for the basic split 4D-Var algorithms: I4DVAR_SPLIT, RBL4DVAR_SPLIT, and R4DVAR_SPLIT. Now, we get identical solutions when comparing unsplit and split solutions with the WC13 test case.
The split strategy is scripted to run with different executables in the elementary background, increment, and analysis 4D-Var phases, as mentioned in src:ticket:862. The modules i4dvar.F, rbl4dvar.F, and r4dvar.F have additional code activated with the internal CPP option SPLIT_4DVAR to load values to several variables which are assigned independently in other phases or are in memory in the unsplit algorithm.
- The minimization solvers congrad.F, cgradinet.F, and rpcg_lanczos.F now include routines cg_read_congrad, cg_read_gradient, and cg_read_rpcg, respectively, to read previous outer loop variables from the MODNAME (DAV structure), which will be on memory in the unsplit algorithm. It will come handy when re-starting 4D-Var in the future. This capability is straight forward in the split drivers. Such drivers (split_i4dvar_ocean.h, split_rbl4dvar_ocean.h, and split_r4dvar_ocean.h) will be released in the future.
- The rpcg_lanczos solver is now inside of a module for strong type checking of routine arguments.
- The routine wrt_impulse now reports statistics including the checksum to facilitate comparison of unsplit and split algorithms. For example, we can have:
Converting Convolved Adjoint Trajectory to Impulses: Outer = 001 Inner = 004 WRT_IMPULSE - processing convolved adjoint impulses, records: 1 to 2 file: wc13_roms_tlf_20040103.nc - zeta (Rec = 2 Min = -1.30850216E-02 Max = 1.55209772E-02 CheckSum = -1441498552) - u (Rec = 2 Min = -5.60027244E+01 Max = 5.89141275E+01 CheckSum = -1370629773) - v (Rec = 2 Min = -6.30660677E+01 Max = 8.89201837E+01 CheckSum = -1908057756) - temp (Rec = 2 Min = -3.88886922E+01 Max = 1.61558954E+01 CheckSum = -2119489230) - salt (Rec = 2 Min = -8.91395412E+02 Max = 3.81362699E+02 CheckSum = 310816551) - zeta (Rec = 1 Min = 0.00000000E+00 Max = 0.00000000E+00 CheckSum = 14278099) - u (Rec = 1 Min = -1.61792544E+01 Max = 1.64542330E+01 CheckSum = 919474362) - v (Rec = 1 Min = -1.44458820E+01 Max = 1.80490759E+01 CheckSum = 661583172) - temp (Rec = 1 Min = -2.87144548E+01 Max = 1.04972849E+01 CheckSum = -500278367) - salt (Rec = 1 Min = -3.36527609E+02 Max = 3.27248419E+02 CheckSum = -205855439)
- The initial conditions for the next R4DVAR data assimilation cycle is now written in the DAINAME NetCDF file. Here, we use the last record of the representer model (RPM) for the final outer loop instead of the nonlinear model:
DO ng=1,Ngrids LdefDAI(ng)=.TRUE. CALL def_dai (ng) IF (FoundError(exit_flag, NoError, __LINE__, & & __FILE__)) RETURN ! WRITE (TLM(ng)%name,10) TRIM(FWD(ng)%head), Nouter 10 FORMAT (a,'_outer',i0,'.nc') lstr=LEN_TRIM(TLM(ng)%name) TLM(ng)%base=TLM(ng)%name(1:lstr-3) IF (TLM(ng)%Nfiles.gt.1) THEN Nfiles=TLM(ng)%Nfiles DO ifile=1,Nfiles WRITE (suffix,"('_',i4.4,'.nc')") ifile TLM(ng)%files(ifile)=TRIM(TLM(ng)%base)//TRIM(suffix) END DO TLM(ng)%name=TRIM(TLM(ng)%files(Nfiles)) ELSE TLM(ng)%files(1)=TRIM(TLM(ng)%name) END IF ! CALL netcdf_get_dim (ng, iRPM, TLM(ng)%name, & & DimName = 'ocean_time', & & DimSize = InpRec) IF (FoundError(exit_flag, NoError, __LINE__, & & __FILE__)) RETURN Tindex=1 CALL get_state (ng, iRPM, 1, TLM(ng)%name, InpRec, Tindex) IF (FoundError(exit_flag, NoError, __LINE__, & & __FILE__)) RETURN ! KOUT=Tindex NOUT=Tindex CALL wrt_dai (ng, tile) IF (FoundError(exit_flag, NoError, __LINE__, & & __FILE__)) RETURN END DOMany thanks to Andy Moore for the suggestion.
- Added new variable NLmodel_final to MODNAME NetCDF file containing the final values of the nonlinear model solution interpolated at the observation locations in a 4D-Var cycle:
double NLmodel_final(datum) ; NLmodel_final:long_name = "final nonlinear model at observation locations" ; NLmodel_final:units = "state variable units" ; NLmodel_final:_FillValue = 1.e+37 ;It is useful to have this variable for processing output MODNAME NetCDF file in ERDDAP when computing the Desroziers et al. (2005) 4D-Var diagnostics. Many thanks to John Wilkin for suggesting this additional variable.
WARNING: The file varinfo.dat was modified to include metadata for NLmodel_final:
'NLmodel_final' ! Output 'final nonlinear model at observation locations' 'state variable units' 'nulvar' 'datum' 'idNLmf' 'nulvar' 1.0d0
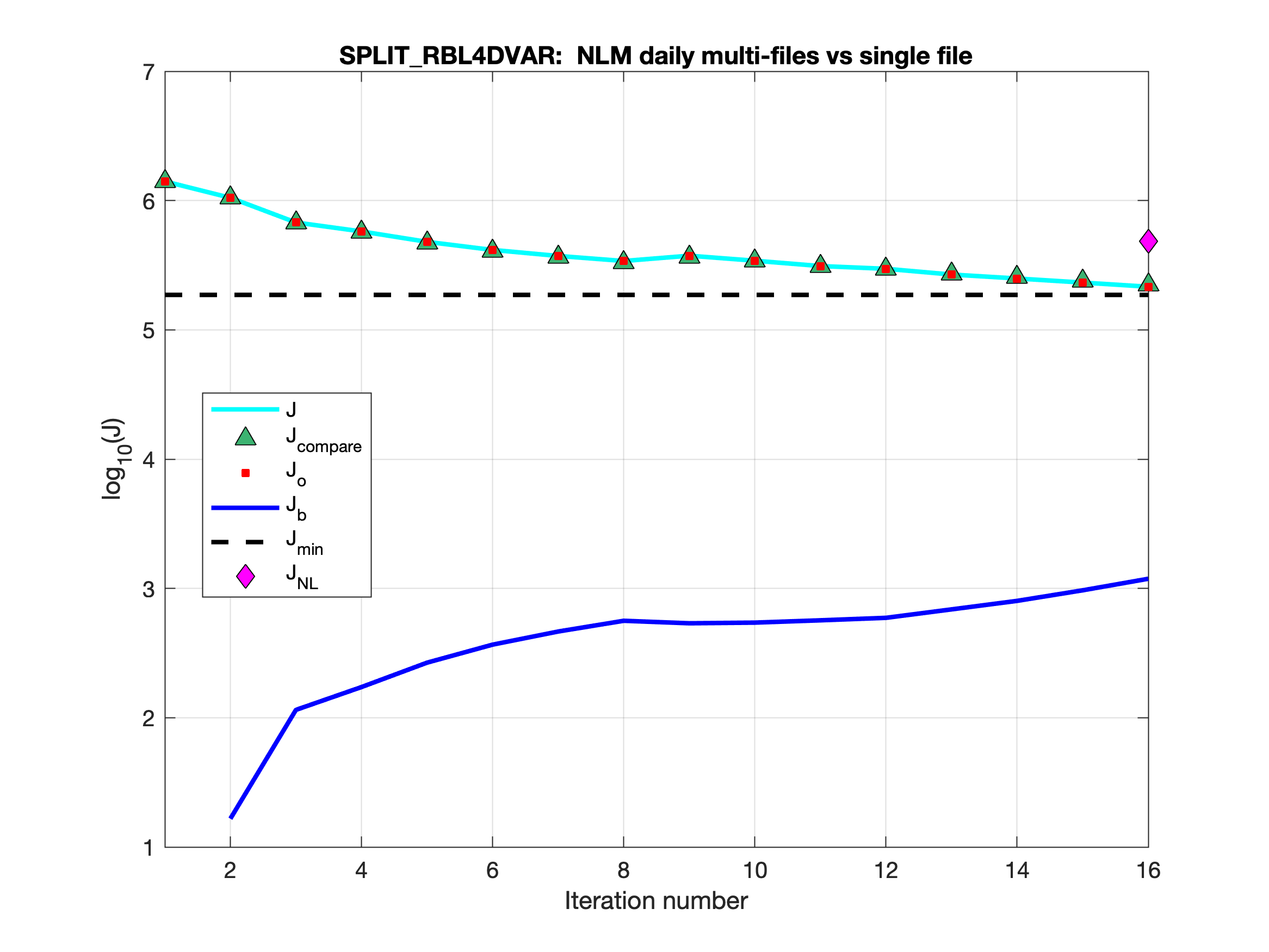
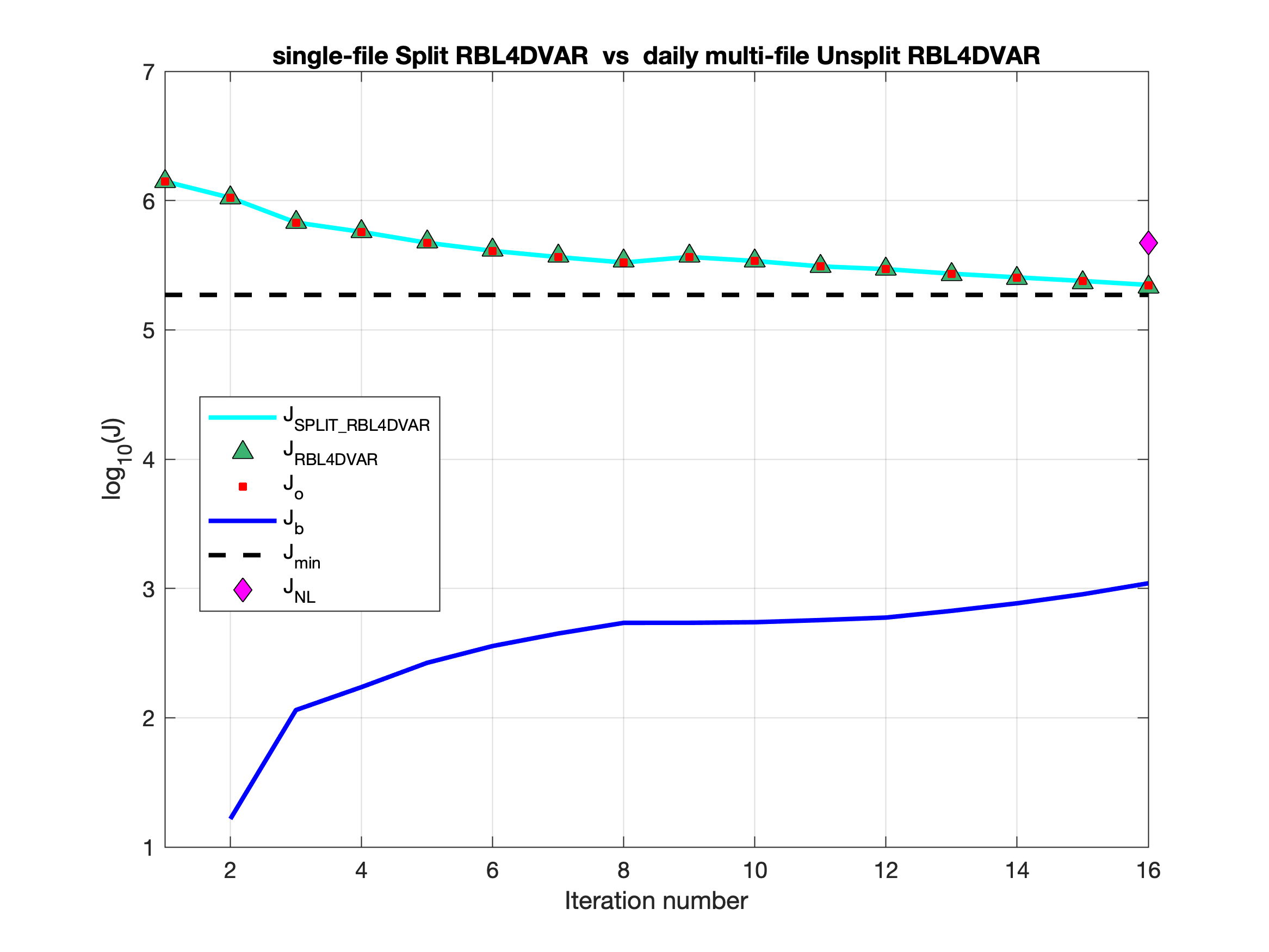
- The nonlinear solution can be split into multi-file in 4D-Var applications to avoid getting huge files. See src:ticket:825 for detailed information. I have been testing this capability for a while and I found out that sometimes the solutions are not bit-by-bit identical. After some testing, I came to the conclusion that the differences are due to roundoff, and the 4D-Var convergence and solution are the same. For example, in our US West Coast DOPPIO application, we get the following cost functions convergence (Nouter=2 and Ninner=8):
The above figure compares two Split RBL4D-Var runs showing the total cost function for the NLM multi-file trajectory case (cyan line) versus the NLM single-file trajectory case (green triangles).
The above figure compares two runs showing the total cost function for Split RBL4D-Var with NLM single-file trajectory (cyan line) versus Unsplit RBLD-Var with multi-file NLM trajectory (green triangles).
Corrected Bug:
Corrected a multi-file bug in output.F, tl_output.F, rp_output.F, and ad_output.F for all ROMS output files in restart applications. For example in output.F, the counter HIS(ng)%load needs reset to zero in restart applications:
IF ((nrrec(ng).ne.0).and.(iic(ng).eq.ntstart(ng))) THEN
IF ((iic(ng)-1).eq.idefHIS(ng)) THEN
HIS(ng)%load=0 ! restart, reset counter
Ldefine=.FALSE. ! finished file, delay
ELSE ! creation of next file
Ldefine=.TRUE.
NewFile=.FALSE. ! unfinished file, inquire
END IF ! content for appending
idefHIS(ng)=idefHIS(ng)+nHIS(ng) ! restart offset
ELSE IF ((iic(ng)-1).eq.idefHIS(ng)) THEN
...
END IF
The same needs to be corrected for the AVG(ng), ADM(ng), QCK(ng), and TLM(ng) structures field load. Many thanks to Julia Levin for bringing this to my attention.
Change History (2)
comment:1 by , 6 years ago
| Description: | modified (diff) |
|---|---|
| Resolution: | → Done |
| Status: | new → closed |

There is still a problem in restart with the multi-file option when MOD(nHIS(ng),ndefHIS(ng)).ne.0. The Nfiles dimension parameter in the structure is exceeded during the creation of the last multi-file. We need to have different logic in read_phypar.F:
DO ng=1,Ngrids IF ((nHIS(ng).gt.0).and.(ndefHIS(ng).gt.0)) THEN OutFiles=ntimes(ng)/ndefHIS(ng) IF ((nHIS(ng).eq.ndefHIS(ng)).or. & & (MOD(ntimes(ng),ndefHIS(ng)).ge.nHIS(ng))) THEN OutFiles=Outfiles+1 END IF CALL edit_file_struct (ng, OutFiles, HIS) END IF ... END DOI coded some safeguard in output.F:
IF (Ldefine) THEN ! create new file or IF (iic(ng).eq.ntstart(ng)) THEN ! inquire existing file HIS(ng)%load=0 ! reset filename counter END IF ifile=(iic(ng)-1)/ndefHIS(ng)+1 ! next filename suffix HIS(ng)%load=HIS(ng)%load+1 IF (HIS(ng)%load.gt.HIS(ng)%Nfiles) THEN IF (Master) THEN WRITE (stdout,10) 'HIS(ng)%load = ', HIS(ng)%load, & & HIS(ng)%Nfiles, TRIM(HIS(ng)%base), & & ifile END IF exit_flag=4 IF (FoundError(exit_flag, NoError, __LINE__, & & __FILE__)) RETURN END IF Fcount=HIS(ng)%load HIS(ng)%Nrec(Fcount)=0 IF (Master) THEN WRITE (HIS(ng)%name,20) TRIM(HIS(ng)%base), ifile END IF ... END IFIf the Nfiles dimension is exceeded, we get the following error:
OUTPUT - multi-file counter HIS(ng)%load = 4, is greater than Nfiles = 3 dimension in structure when creating next file: doppio_his_5308_0007.nc Incorrect OutFiles logic in 'read_phypar'. Found Error: 04 Line: 122 Source: ROMS/Nonlinear/output.F Found Error: 04 Line: 518 Source: ROMS/Nonlinear/main3d.F Found Error: 04 Line: 300 Source: ROMS/Drivers/nl_ocean.h