Code: Select all
iSphy small phytoplankton biomass, nanophytoplankton
iLphy large phytoplankton biomass, diatoms
iSzoo small zooplankton biomass, microzooplankton (ciliates)
iLzoo large zooplankton biomass, mesozooplankton (copepods)
iPzoo predator zooplankton biomass (euphausiids, etc)
iNO3_ nitrate concentration, NO3
iNH4_ ammonium concentration, NH4
iPON_ particulate organic nitrogen
iDON_ dissolved organic nitrogen
iSiOH silicate concentration, Si(OH)4 (silicic acid)
iopal particulate organic silica
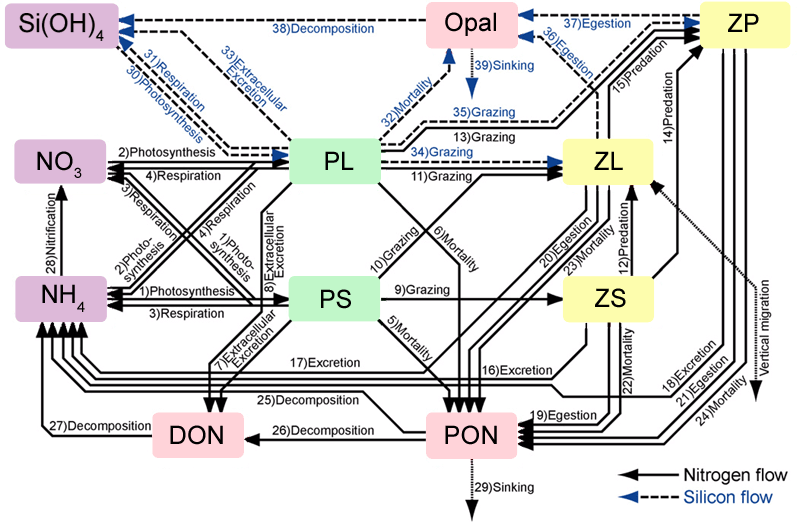
The basic reference for this model is Kishi et al. (2007)
I coded this model from scratch using the same numerical scheme of other existing biological models in ROMS. The compartment being consumed is treated implicit to guarantee conservation, a positive definite and unconditionally stable algorithm. The model can be activated with the NEMURO cpp option.
Currently, it has three formulations for the grazing terms:
- Explit Ivlev grazing formulation, original code. Activated with option IVLEV_EXPLICIT.
- Holling-type s-shape grazing curve with uses a quadratic term on zooplankton. It is known to be numerically more stable and allows an implicit formulation. Activated with option HOLLING_GRAZING.
- Implicit Ivlev grazing formulation, default option. The implicit algorithm is possible after some algebraic transformation and approximation with Taylor series. Many thanks to Chris Edwards for his help in this formulation. This formulation does not work well yet. The model blows-up sometimes. It is recommended to use either IVLEV_EXPLICIT or HOLLING_GRAZING
This model is available in the ROMS svn repository (revision 82 or higher). The input script nemuro.in can be found in ROMS/External. There is also a copy in User/External. As in all ecosystem models in ROMS, the compartment concentrations are in millimoles/m3. I added a simple initialization in ana_biology.h which is located in ROMS/Functionals with a copy in User/Functionals.
Warning: the model is not fully tested