Recently while configuring river locations I discovered that ROMS wasn't reading
masks on my NetCDF grid file correctly; not as I wrote them, nor as they appeared
when viewing the grid input file with ncView. Here's an example of what ROMS read.
The central cell is the target and surrounding cells are shown for context:
rmask umask vmask
1.1.1.1.1. 1.1.1.1.1. 1.1.0.0.1.
1.1.1.1.1. 1.1.1.1.1. 1.0.0.0.0.
1.1.0.0.1. 1.1.1.0.0. 0.0.0.0.0.
1.0.0.0.0. 1.1.0.0.0. 0.0.0.0.0.
0.0.0.0.0. 0.0.0.0.0. 0.0.0.0.0.
and in wrt_his.F wrote:
vmask_io
1.1.0.0.1.
1.0.0.0.0.
0.0.0.0.0.
0.0.0.0.0.
0.0.0.0.0.
Notice that vmask is offset one extra cell upward.
After modifying nf_read2d.F, lines 657 to 670 to change reading offsets:
Imin=IOBOUNDS(ng)%ILB_u-1
Imax=IOBOUNDS(ng)%IUB_u-1
Jmin=IOBOUNDS(ng)%JLB_u
Jmax=IOBOUNDS(ng)%JUB_u
CASE (v2dvar)
Imin=IOBOUNDS(ng)%ILB_v
Imax=IOBOUNDS(ng)%IUB_v
Jmin=IOBOUNDS(ng)%JLB_v-1
Jmax=IOBOUNDS(ng)%JUB_v-1
CASE DEFAULT
Imin=IOBOUNDS(ng)%ILB_rho-1
Imax=IOBOUNDS(ng)%IUB_rho-1
Jmin=IOBOUNDS(ng)%JLB_rho-1
Jmax=IOBOUNDS(ng)%JUB_rho-1
ROMS read:
rmask umask vmask
1.1.1.1.1. 1.1.1.1.1. 1.1.1.1.1.
1.1.1.1.1. 1.1.1.1.1. 1.1.0.0.1.
1.1.0.0.1. 1.1.0.0.0. 1.0.0.0.0.
1.0.0.0.0. 1.0.0.0.0. 0.0.0.0.0.
0.0.0.0.0. 0.0.0.0.0. 0.0.0.0.0.
and in wrt_his.F wrote:
vmask_io
1.1.1.1.1.
1.1.0.0.1.
1.0.0.0.0.
0.0.0.0.0.
0.0.0.0.0.
Before this change I couldn't coax ROMS to model more than about 30 minutes of ocean time,
nor would it do reasonable river flow.
This may be a NetCDF bug due to differences between files produced with C++ programs and
Fortran programs. I write my input files with C++.
rivers and masking read from grid file
Re: rivers and masking read from grid file
Your C++ program might not be taking into account how the edges occur. In the Fortran, the vmask starts at j=1, but in the netcdf file (or in C++), that is considered to be j=0. Likewise, umask (and other u variables) start at i=1 in the Fortran.
Since the rho mask determines all the rest, why not have ROMS read just that, then compute the rest? It's already recomputing the phi mask.
Since the rho mask determines all the rest, why not have ROMS read just that, then compute the rest? It's already recomputing the phi mask.
-
rubash
Re: rivers and masking read from grid file
The model region is a fully enclosed lake and I left a margin on all sides so I wouldn't have to worry about the outer boundaries, so boundary edges are not likely to be a problem. I think I have the indexing difference between C++ and Fortran right because the mask_rho points are all in the right places.
Kate wrote:
"Since the rho mask determines all the rest, why not have ROMS read just that, then compute the rest? It's already recomputing the phi mask"
How can I do that? I tried leaving the offending masks out but ROMS said:
GET_GRID - unable to find grid variable: mask_u in grid NetCDF file: ocean_grd.nc
Kate wrote:
"Since the rho mask determines all the rest, why not have ROMS read just that, then compute the rest? It's already recomputing the phi mask"
How can I do that? I tried leaving the offending masks out but ROMS said:
GET_GRID - unable to find grid variable: mask_u in grid NetCDF file: ocean_grd.nc
Re: rivers and masking read from grid file
Sorry, it would have to be a policy change from Hernan unless you felt like hacking it in yourself.
What I meant about the indices is that the rho mask starts at 0 in netCDf, Fortran, C++, consistently. The other masks start at 1 in the Fortran, in at least one dimension. You have to be consistent with what ROMS wants when building new tools. If you got things to work with an offset, that's probably what ROMS wants. This figure describes what ROMS is looking for from its input fields:
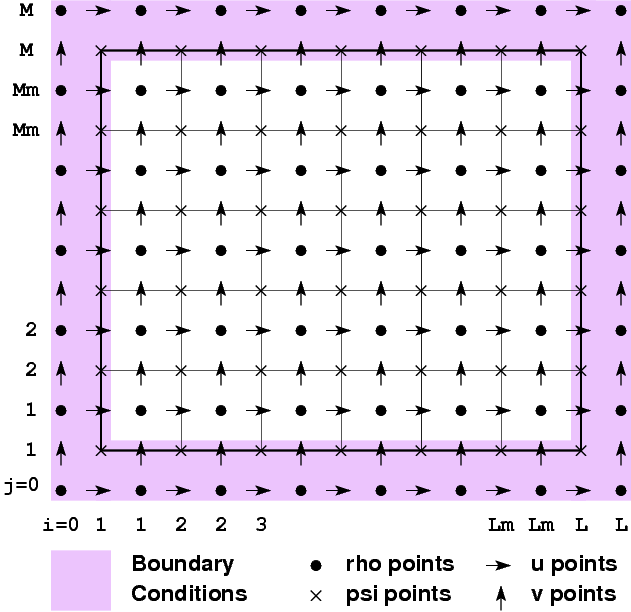
What I meant about the indices is that the rho mask starts at 0 in netCDf, Fortran, C++, consistently. The other masks start at 1 in the Fortran, in at least one dimension. You have to be consistent with what ROMS wants when building new tools. If you got things to work with an offset, that's probably what ROMS wants. This figure describes what ROMS is looking for from its input fields:

-
rubash
Re: rivers and masking read from grid file
Thanks Kate; after pondering the diagram I transferred the changes I made from nf_read2d.f to the grid creating program that I had written and everything works satisfactorily.
I predict that ROMS' filing and reading offset arrangements will trip other ROMS users who use grid input files with masks and rivers and tracers.
I predict that ROMS' filing and reading offset arrangements will trip other ROMS users who use grid input files with masks and rivers and tracers.