Phase III, Major Overhaul of ROMS Kernel to Complete Nesting
The ROMS algorithms where
Phase III was particularly difficult and it took long time to finish. I know that some of you have been patiently waiting for it. I think that the wait was worth it and ROMS nesting has tremendous capabilities and potential. I rewrote nesting.F several times because I was looking for a very elegant and generic algorithm for nesting in ROMS. The most difficult part was the Nested Grid Connectivity (NGC) for any generic application. This is all done outside of ROMS and provided as in a new input NetCDF file (NGCname).
The nesting algorithms took long to be developed in this version ROMS. Many thanks to John Warner for helping us to develop and test this capability.
The idea for nesting evolved with time and we became more ambitious as you can see in the technical documentation in
The nesting in ROMS is always two-way. There is a duality for each nested grid: data donor in one contact region and data receiver in its conjugate contact region. Thus, we prefer to use the donor and receiver categories instead of the usual parent and child descriptions found in the literature.
What Is New:
- Rewrote completely module mod_nesting.F. It now contains several structures to process nesting contact regions and contact points:
- Nested Grid Connectivity structure, type NGC:
This structure is used to store all the connectivity information between nested grids. It is used extensible when processing contact region points between data donor and data receiver grids. This information is processed outside of ROMS and read from input NGCname NetCDF file for functionality and efficiency. In nested grids, the points in the contact region are interpolated from the data donor grid cell containing the contact point using the following conventions at the horizontal location in the receiver grid (Irg, Jrg) and donor grid cell (Idg, Jdg):
Code: Select all
integer :: Ncontact ! total number of contact regions ! TYPE T_NGC logical :: coincident ! coincident donor/receiver, p=q=0 logical :: interpolate ! perform vertical interpolation integer :: donor_grid ! data donor grid number integer :: receiver_grid ! data receiver grid number integer :: Npoints ! number of points in contact region integer, pointer :: Irg(:) ! receiver grid, I-contact point integer, pointer :: Jrg(:) ! receiver grid, J-contact point integer, pointer :: Idg(:) ! donor grid, cell I-left index integer, pointer :: Jdg(:) ! donor grid, cell J-bottom index # ifdef SOLVE3D integer, pointer :: Kdg(:,:) ! donor grid, cell K-index # endif real(r8), pointer :: Hweight(:,:) ! horizontal weights # ifdef SOLVE3D real(r8), pointer :: Vweight(:,:,:) ! vertical weights # endif END TYPE T_NGC TYPE (T_NGC), allocatable :: Rcontact(:) ! RHO-points, [Ncontact] TYPE (T_NGC), allocatable :: Ucontact(:) ! U-points, [Ncontact] TYPE (T_NGC), allocatable :: Vcontact(:) ! V-points, [Ncontact]
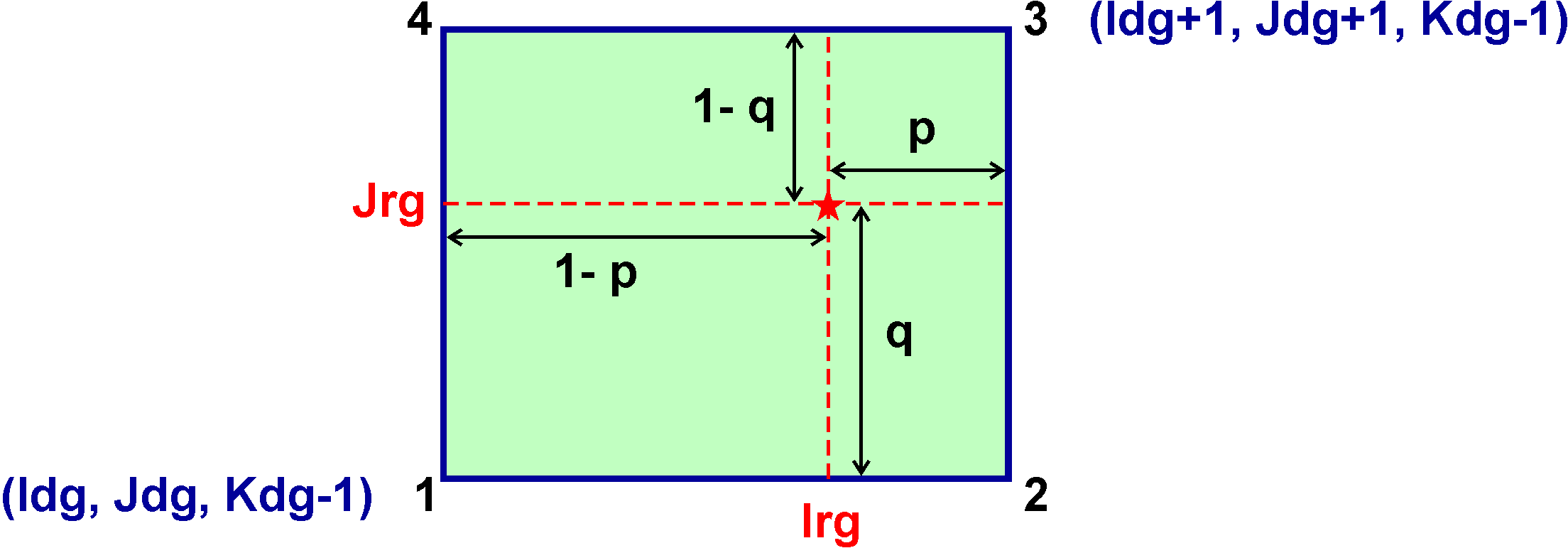
Here, the suffixes dg and rg are for the donor and receiver grid, respectively. The horizontal interpolation weights Hweight(1:4,m) for contact point m are computed outside of ROMS and read from the NGCname NetCDF file. If coincident contact points between data donor and data receiver grids, p = q = 0. Then, Hweight(1,m)=1 and Hweight(2:3,m)=0. - Boundary Contact Point structure, type BCP:
This structure is allocated as [4,Ncontact]. Currently, it is used in refinement grids where the coarser (donor) and finer (receiver) grids have coincident physical boundaries but with different I- and J-indices. The variable C2Bindex (Coarse to Boundary index) is used to tell us which contact points in the Ucontact and Vcontact structures are located at the physical boundary of the relevant nested grid. For example at the boundary edge of a grid with contact region cr, we can get the mapping between contact point m and grid physical boundary edge index i or j as:
Code: Select all
TYPE T_BCP integer :: spv ! fill value, unwanted index integer :: Ibmin ! viable minimum Ib integer :: Ibmax ! viable maximum Ib integer :: Jbmin ! viable minimum Jb integer :: Jbmax ! viable maximum Jb integer, pointer :: Ib(:) ! I-boundary index integer, pointer :: Jb(:) ! J-boundary index integer, pointer :: C2Bindex(:) ! contact to boundary index real(r8), pointer :: Tflux(:,:,:) ! Perimeter tracer flux END TYPE T_BCP TYPE (T_BCP), allocatable :: BRY_CONTACT(:,:) ! [4,Ncontact]
Code: Select all
m = BRY_CONTACT(iwest, cr) % C2Bindex(j) m = BRY_CONTACT(isouth,cr) % C2Bindex(i) m = BRY_CONTACT(ieast, cr) % C2Bindex(j) m = BRY_CONTACT(inorth,cr) % C2Bindex(i) - Nested Grids Metrics structure, type NGM:
In some nesting configurations, there are contact points outside of the regular (physical) nested grid domain. That is, such contact points are located in the extended numerical regions. These metrics values need to be computed when designing and generating the applications grids.Code: Select all
TYPE T_NGM real(r8), pointer :: angler(:) ! angle between XI and EAST real(r8), pointer :: dndx(:) ! d(1/pn)/d(XI) real(r8), pointer :: dmde(:) ! d(1/pm)/d(ETA) real(r8), pointer :: f(:) ! Coriolis parameter real(r8), pointer :: h(:) ! bathymetry real(r8), pointer :: rmask(:) ! land/sea RHO-mask real(r8), pointer :: umask(:) ! land/sea U-mask real(r8), pointer :: vmask(:) ! land/sea V-mask real(r8), pointer :: pm(:) ! XI-coordinate metric real(r8), pointer :: pn(:) ! ETA-coordinate metric real(r8), pointer :: Xr(:) ! X RHO-coordinate (m or degrees) real(r8), pointer :: Yr(:) ! Y RHO-coordinate (m or degrees) real(r8), pointer :: Xu(:) ! X U-coordinate (m or degrees) real(r8), pointer :: Yu(:) ! Y U-coordinate (m or degrees) real(r8), pointer :: Xv(:) ! X V-coordinate (m or degrees) real(r8), pointer :: Yv(:) ! Y V-coordinate (m or degrees) END TYPE T_NGM TYPE (T_NGM), allocatable :: CONTACT_METRIC(:) ! [Ncontact] In such cases, it is recommended to build an intermediary fine resolution grid encompassing the study area firsts and extract/sample all the ROMS application nested grids (composite and refinement) from it. This would give a better handle on volume conservation, bathymetry representation, land/sea masking and other grid topology issues. These metrics are read from NGCname NetCDF and packed in this structure. It is very tricky to load these values directly into the global grid metrics arrays because of parallelization.
In such cases, it is recommended to build an intermediary fine resolution grid encompassing the study area firsts and extract/sample all the ROMS application nested grids (composite and refinement) from it. This would give a better handle on volume conservation, bathymetry representation, land/sea masking and other grid topology issues. These metrics are read from NGCname NetCDF and packed in this structure. It is very tricky to load these values directly into the global grid metrics arrays because of parallelization. - Composite Grids structure, type COMPOSITE:
It contains the donor grid data at the receiver grid contact points. The donor grid data is extracted for the cell containing the contact point: 4 horizontal values to facilitate spatial interpolation.Code: Select all
TYPE T_COMPOSITE real(r8), pointer :: bustr(:,:) ! [4,Npoints] real(r8), pointer :: bvstr(:,:) ! [4,Npoints) real(r8), pointer :: ubar(:,:,:) ! [4,Npoints,2] real(r8), pointer :: vbar(:,:,:) ! [4,Npoints,2] real(r8), pointer :: zeta(:,:,:) ! [4,Npoints,2] real(r8), pointer :: rzeta(:,:) ! [4,Npoints] # ifdef SOLVE3D real(r8), pointer :: DU_avg1(:,:) ! [4,Npoints] real(r8), pointer :: DV_avg1(:,:) ! [4,Npoints] real(r8), pointer :: Zt_avg1(:,:) ! [4,Npoints] real(r8), pointer :: u(:,:,:) ! [4,k,Npoints] real(r8), pointer :: v(:,:,:) ! [4,k,Npoints] real(r8), pointer :: Huon(:,:,:) ! [4,k,Npoints] real(r8), pointer :: Hvom(:,:,:) ! [4,k,Npoints] real(r8), pointer :: t(:,:,:,:) ! [4,k,Npoints,itrc] # endif END TYPE T_COMPOSITE TYPE (T_COMPOSITE), allocatable :: COMPOSITE(:) ! [Ncontact] - Refinement Grids structure, type REFINED:
It contains the coarser grid data at the finer grid contact points. The finer grid data is extracted for the cell containing the contact point: 4 horizontal values and 2 time records (t1:t2) to facilitate the space-time linear interpolation.Code: Select all
TYPE T_REFINED real(r8), pointer :: ubar(:,:,:) ! [4,Npoints,t1:t2] real(r8), pointer :: vbar(:,:,:) ! [4,Npoints,t1:t2] real(r8), pointer :: zeta(:,:,:) ! [4,Npoints,t1:t2] real(r8), pointer :: DU_avg2(:,:,:) ! [4,Npoints,t1:t2] real(r8), pointer :: DV_avg2(:,:,:) ! [4,Npoints,t1:t2] # ifdef SOLVE3D real(r8), pointer :: u(:,:,:,:) ! [4,k,Npoints,t1:t2] real(r8), pointer :: v(:,:,:,:) ! [4,k,Npoints,t1:t2] real(r8), pointer :: t(:,:,:,:,:) ! [4,k,Npoints,t1:t2,itrc] # endif END TYPE T_REFINED ! TYPE (T_REFINED), allocatable :: REFINED(:) ! [Ncontact]
- Nested Grid Connectivity structure, type NGC:
- Added new routine set_contact.F to read and process nested grids connectivity data from NGCname NetCDF:
All the nested data is packed in a long vector dimensioned Ndatum. Several indices (NstrR, NendR, NstrU, NendU, NstrV, and NendV) are available to unpack the data in the Rcontact, Ucontact, and Vcontact NGC-type structures for contact point information at RHO-, U-, and V-points. Although NetCDF-4 supports derived type structures via the HDF5 library, a simpler Metadata model is chosen for interoperability. Notice that the NGCname NetCDF file also contains the values for the various grid metrics needed in ROMS numerical kernel. Only the horizontal interpolation weights (Hweight)are provided here since they are always the same (time independent) for a particular configuration. The vertical interpolation weights are computed internally in ROMS at each time step because of the time dependency of the vertical coordinates to the evolution of the free-surface.Code: Select all
netcdf dogbone_ngc_refined { dimensions: Ngrids = 2 ; Ncontact = 2 ; Nweights = 4 ; datum = 1675 ; variables: int spherical ; spherical:long_name = "grid type logical switch" ; spherical:flag_values = 0, 1 ; spherical:flag_meanings = "Cartesian spherical" ; int Lm(Ngrids) ; Lm:long_name = "number of interior RHO-points in the I-direction" ; int Mm(Ngrids) ; Mm:long_name = "number of interior RHO-points in the J-direction" ; int coincident(Ngrids) ; coincident:long_name = "coincident donor and receiver grids logical switch" ; coincident:flag_values = 0, 1 ; coincident:flag_meanings = "false true" ; int composite(Ngrids) ; composite:long_name = "composite grid type logical switch" ; composite:flag_values = 0, 1 ; composite:flag_meanings = "false true" ; int mosaic(Ngrids) ; mosaic:long_name = "mosaic grid type logical switch" ; mosaic:flag_values = 0, 1 ; mosaic:flag_meanings = "false true" ; int refinement(Ngrids) ; refinement:long_name = "refinement grid type logical switch" ; refinement:flag_values = 0, 1 ; refinement:flag_meanings = "false true" ; int refine_factor(Ngrids) ; refine_factor:long_name = "refinement factor from donor grid" ; int interpolate(Ncontact) ; interpolate:long_name = "vertical interpolation at contact points logical switch" ; interpolate:flag_values = 0, 1 ; interpolate:flag_meanings = "false true" ; int donor_grid(Ncontact) ; donor_grid:long_name = "data donor grid number" ; int receiver_grid(Ncontact) ; receiver_grid:long_name = "data receiver grid number" ; int NstrR(Ncontact) ; NstrR:long_name = "starting contact RHO-point index in data vector" ; int NendR(Ncontact) ; NendR:long_name = "ending contact RHO-point index in data vector" ; int NstrU(Ncontact) ; NstrU:long_name = "starting contact U-point index in data vector" ; int NendU(Ncontact) ; NendU:long_name = "ending contact U-point index in data vector" ; int NstrV(Ncontact) ; NstrV:long_name = "starting contact V-point index in data vector" ; int NendV(Ncontact) ; NendV:long_name = "ending contact V-point index in data vector" ; int contact_region(datum) ; contact_region:long_name = "contact region number" ; int on_boundary(datum) ; on_boundary:long_name = "contact point on receiver grid physical boundary" ; on_boundary:flag_values = 0, 1, 2, 3, 4 ; on_boundary:flag_meanings = "other western southern eastern northern" ; int Idg(datum) ; Idg:long_name = "I-left index of donor cell containing contact point" ; int Jdg(datum) ; Jdg:long_name = "J-bottom index of donor cell containing contact point" ; int Irg(datum) ; Irg:long_name = "receiver grid I-index of contact point" ; Irg:coordinates = "Xrg Yrg" ; int Jrg(datum) ; Jrg:long_name = "receiver grid J-index of contact point" ; Jrg:coordinates = "Xrg Yrg" ; double Xrg(datum) ; Xrg:long_name = "X-location of receiver grid contact points" ; Xrg:units = "meter" ; double Yrg(datum) ; Yrg:long_name = "Y-location of receiver grid contact points" ; Yrg:units = "meter" ; double Hweight(datum, Nweights) ; Hweight:long_name = "horizontal interpolation weights" ; Hweight:coordinates = "Xrg Yrg" ; double h(datum) ; h:long_name = "bathymetry at RHO-points" ; h:units = "meter" ; h:coordinates = "Xrg Yrg" ; h:_FillValue = 1.e+37 ; double f(datum) ; f:long_name = "Coriolis parameter at RHO-points" ; f:units = "second-1" ; f:coordinates = "Xrg Yrg" ; f:_FillValue = 1.e+37 ; double pm(datum) ; pm:long_name = "curvilinear coordinate metric in XI" ; pm:units = "meter-1" ; pm:coordinates = "Xrg Yrg" ; pm:_FillValue = 1.e+37 ; double pn(datum) ; pn:long_name = "curvilinear coordinate metric in ETA" ; pn:units = "meter-1" ; pn:coordinates = "Xrg Yrg" ; pn:_FillValue = 1.e+37 ; double dndx(datum) ; dndx:long_name = "XI-derivative of inverse metric factor pn" ; dndx:units = "meter" ; dndx:coordinates = "Xrg Yrg" ; dndx:_FillValue = 1.e+37 ; double dmde(datum) ; dmde:long_name = "ETA-derivative of inverse metric factor pm" ; dmde:units = "meter" ; dmde:coordinates = "Xrg Yrg" ; dmde:_FillValue = 1.e+37 ; double angle(datum) ; angle:long_name = "angle between XI-axis and EAST" ; angle:units = "radians" ; angle:coordinates = "Xrg Yrg" ; angle:_FillValue = 1.e+37 ; double mask(datum) ; mask:long_name = "land-sea mask of contact points" ; mask:flag_values = 0., 1. ; mask:flag_meanings = "land water" ; mask:coordinates = "Xrg Yrg" ; // global attributes: :type = "ROMS Nesting Contact Regions Data" ; :grid_files = " \n", "/Users/arango/ocean/repository/test/dogbone/Data/dogbone_grd_hfull.nc \n", "/Users/arango/ocean/repository/test/dogbone/Data/dogbone_grd_href3.nc" ; :history = "Contact points created by contact.m and written by write_contact.m, Saturday - March 2, 2013 - 8:41:2.664 AM" ; This NetCDF file is created using the Matlab script
This NetCDF file is created using the Matlab script  contact.m. The routine set_contact is called in inp_par after reading the standard input parameters.
contact.m. The routine set_contact is called in inp_par after reading the standard input parameters. - Added new module nesting.F to process nesting capabilities in ROMS. It contains several public and private routines:
- CALL nesting (ng, model, isection), public interface to ROMS time-stepping kernel. This routine is the main driver for nesting. It performs the various nesting operations according to the isection flag with the following values:
Code: Select all
! !----------------------------------------------------------------------- ! Nesting identification index of variables to process. !----------------------------------------------------------------------- ! ! The following identification indices are used in main2d/main3d ! to specify the variables that are processed in each sub-timestep ! section. Negative indices are used in grid refinement whereas ! positive indices are used in composite grids. ! integer, parameter :: ngetD = -3 ! extract donor grid data integer, parameter :: nputD = -2 ! fill contact points integer, parameter :: n2way = -1 ! fine to course coupling ! integer, parameter :: nFSIC = 1 ! free surface initialization integer, parameter :: n2dIC = 2 ! 2D momentum initialization integer, parameter :: n3dIC = 3 ! 3D momentum initialization integer, parameter :: nTVIC = 4 ! tracers initialization integer, parameter :: nbstr = 5 ! bottom stress (bustr,bvstr) integer, parameter :: nrhst = 6 ! RHS terms (tracers) integer, parameter :: nzeta = 7 ! 3D kernel free-surface integer, parameter :: nzwgt = 8 ! 3D vertical weights integer, parameter :: n2dPS = 9 ! 2D engine Predictor Step integer, parameter :: n2dCS = 10 ! 2D engine Corrector Step integer, parameter :: n2dfx = 11 ! time-averaged 2D fluxes integer, parameter :: n3duv = 12 ! 3D momentum and fluxes integer, parameter :: n3dTV = 13 ! 3D tracer variables - CALL bry_fluxes (dg, rg, cr, model, tile, ...), this public routine extracts tracer horizontal advective fluxes (Hz*u*T/n, Hz*v*T/m) at the grid contact boundary (physical domain perimeter). The data source is either the coarse or finer grid and it is call from tracer stepping routine step3d_t. These fluxes are used for in two-way nesting.
- CALL fill_contact (rg, model, tile, cr, ...), this public routine is used during initialization to fill the contact points of a specified grid metric array. We need to have metric data in all the extended computational points of the grid. No attempt is done here to interpolate such values since the are read in set_contact from input contact points NGCname NetCDF file. This routine just unpack data into global arrays and check if all needed values are filled. Notice that during allocation these special metric grid arrays are initialized to spval to avoid resetting those values processed already from the regular Grid NetCDF file. That is, only those contact points outside the physical grid are processed here. This is a good way to check if all the extra numerical points have been processed.
- CALL correct_tracer (ng, ngf, model, tile) , this private routine corrects the tracer values in the coarser grid, ng, at the location of the finer grid, ngf, physical domain perimeter by comparing vertically accumulated horizontal tracer flux (Hz*u*T/n, Hz*v*T/m) in two-way nesting refinement. It uses the horizontal advective fluxes extracted in routine bry_fluxes.
- CALL fine2coarse (ng, model, vtype, tile), this private replaces interior coarser grid data with the refined averaged values: two-way nesting. It is called separated for 2D and 3D state-variables using the vtype flag.
- CALL get_composite (ng, model, isection, tile), this private routine gets the donor grid data required to process the contact points of the current composite grid, ng. It extracts the donor cell points containing each contact point. In composite grids, it is possible to have more than one contact region.
- CALL get_contact2d (dg, model, tile, gtype, svname, ...), this private low level generic routine gets the donor grid data necessary to process the contact points for a 2D state variable. It extracts the donor cell points containing each contact point. It is called repetitively from get_composite and get_refined.
- CALL get_contact3d (dg, model, tile, gtype, svname, ...), this private low level generic routine gets the donor grid data necessary to process the contact points for a 3D state variable. It extracts the donor cell points containing each contact point. It is called repetitively from get_composite and get_refined.
- CALL get_refine (ng, model, tile), this private routine gets the donor grid data required to process the contact points of the current refinement grid. It extracts the donor cell points containing each contact point. The extracted data is stored in two-time rolling records which to allow the temporal interpolation in put_refine.
- CALL put_composite (ng, model, isection, tile), this private routine interpolates composite grid, ng, contact points from donor grid data extracted in routine get_composite.
- put_contact2d (rg, model, tile, gtype, svname, ...), this private low level routine uses extracted donor grid data to spatially interpolate a 2D state variable at the receiver grid, rg, contact points. If the donor and receiver grids are coincident, the spatial weights Hweight(1,:)=1 and Hweight(2:4,:)=0. This routine is called repetitively from put_composite.
- put_contact3d (rg, model, tile, gtype, svname, ...), this private low level routine uses extracted donor grid data to spatially interpolate a 3D state variable at the receiver grid, rg, contact points. If the donor and receiver grids are coincident, the spatial weights Hweight(1,:)=1 and Hweight(2:4,:)=0. This routine is called repetitively from put_composite.
- CALL put_refine (ng, model, tile) , this private routine spatially interpolates composite grid contact, ng, points from donor grid data extracted in routine get_composite.
- put_refine2d (ng, dg, cr, model, tile, ...), this private routine performs a spatial and temporal linear interpolates of the refinement grid, ng, 2D state variables contact points using data from the donor grid. It imposes mass flux from the coarser grid at the finer grid physical boundaries. It is called from routine put_refine.
- put_refine3d (ng, dg, cr, model, tile, ...), this private routine performs a spatial and temporal linear interpolates of the refinement grid, ng, 3D state variables contact points using data from the donor grid. It is called from routine put_refine.
- CALL z_weights (ng, model, tile), this private routine determines the vertical indices (Kdg) and vertical interpolation weights (Vweight) associated with the time-evolving depths, which are needed to process 3D fields in the contact region.
- CALL nesting (ng, model, isection), public interface to ROMS time-stepping kernel. This routine is the main driver for nesting. It performs the various nesting operations according to the isection flag with the following values:
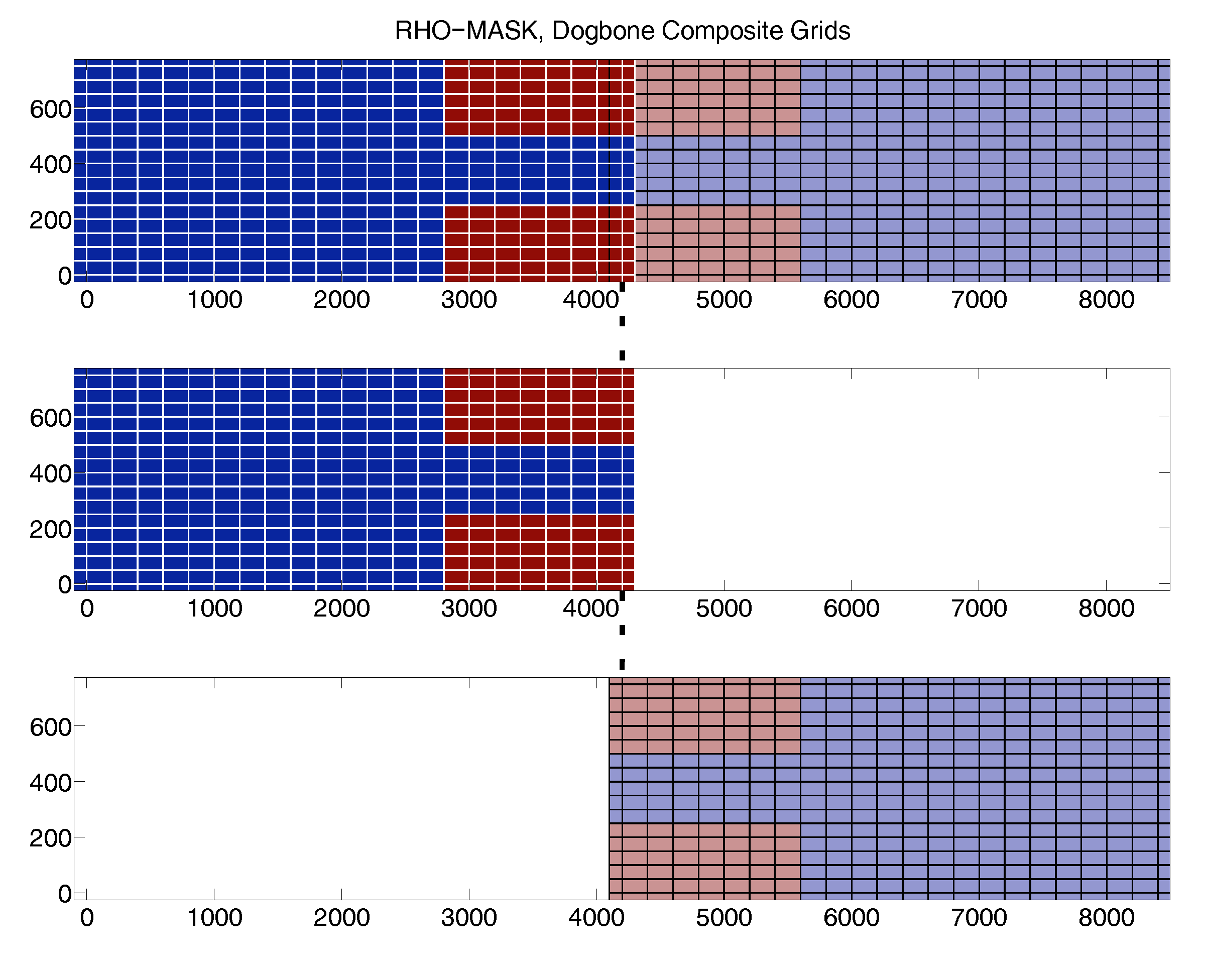
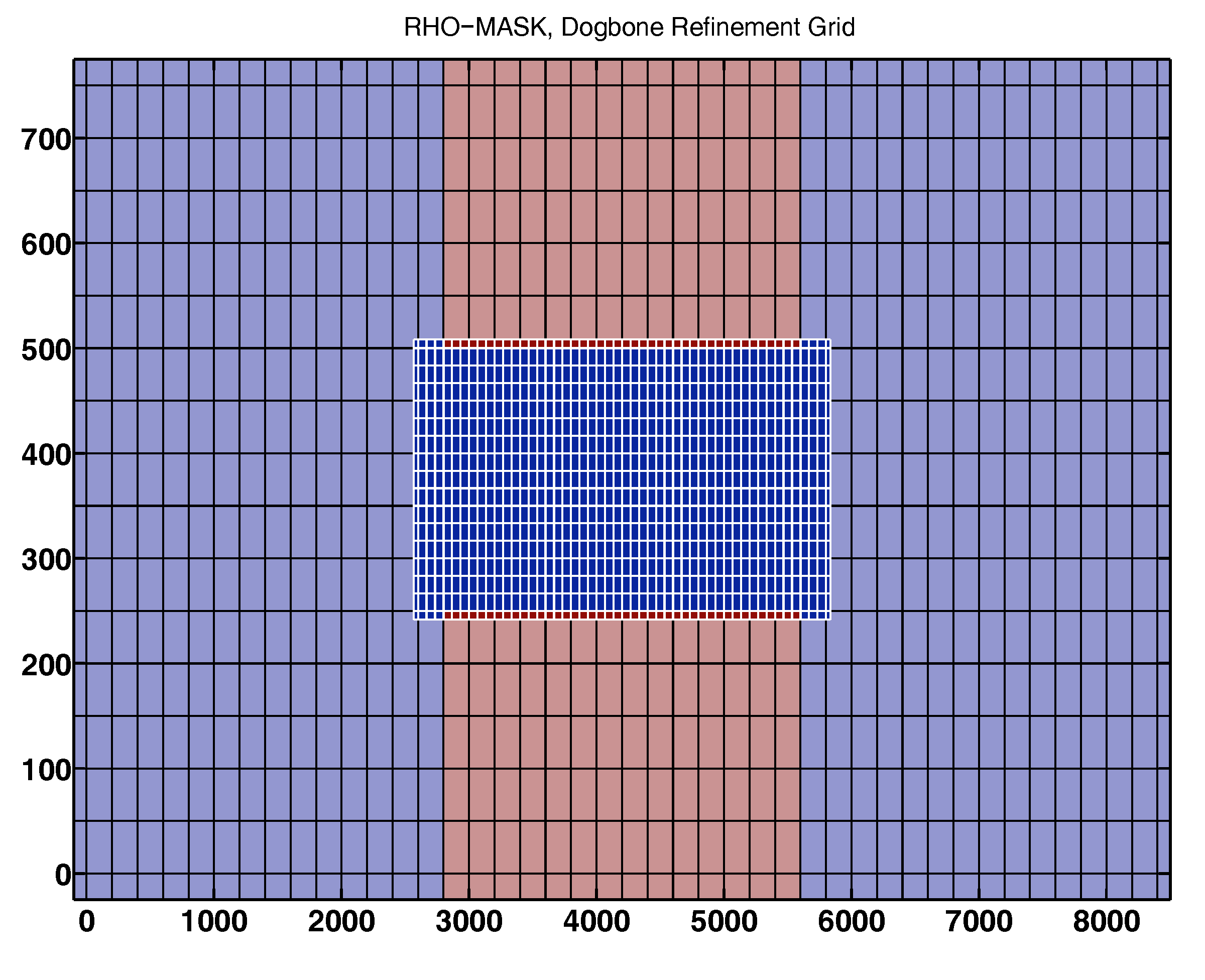
Test Case: DOGBONE